| Size | Volume | Purity |
|---|---|---|
| <25 kB | 10 µL | Unpurified |
Prepare your sample.
We accept RCA (rolling circle amplification) products by popular demand from customers, but make no guarantee of the quality of results. Please note, RCA tends to make chimeric DNA, which can cause errors in our assembly algorithms.
RCA samples must be < 25 kb. Submit at least 10 μL of each unpurified RCA reaction in 200 μl strip tubes.
Label your tubes or plates.
- Set up your order online and receive your unique 6-character order ID.
- Write the first 2 characters of your order ID on the 1st tube of each strip
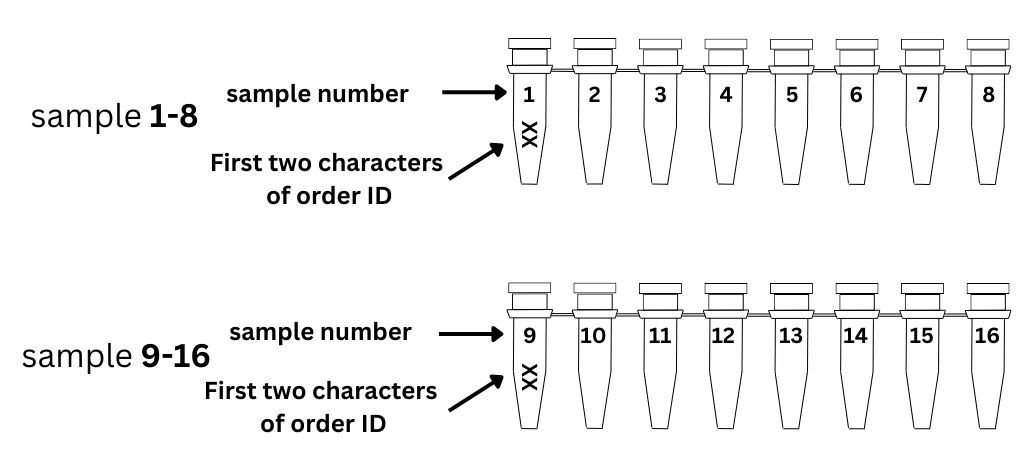
- Set up your order online and receive your unique 6-character order ID. On the order page, please check the “Is this a plate submission?” checkbox, then select if your plate is numbered sequentially by column or by row.
- Label the front edge of the plate with the full 6-character order ID.
- If you are using column numbering, label the first column (i.e., samples 1 to 8). If you are using row numbering, label the first row (i.e., samples 1 to 12).
Load and package your sample.
Add the correct volume of your sample(s) from the table above. Make sure to seal and package your samples well to make sure they arrive safely for sequencing!
- Ensure caps fit snugly and close them firmly. Do not use brands of strip tubes where the caps can easily pop open during shipping.
- Place the labeled tubes into a small protective vessel (such as a Falcon tube, a small box, or bubble wrap) to protect them during shipping. Why?
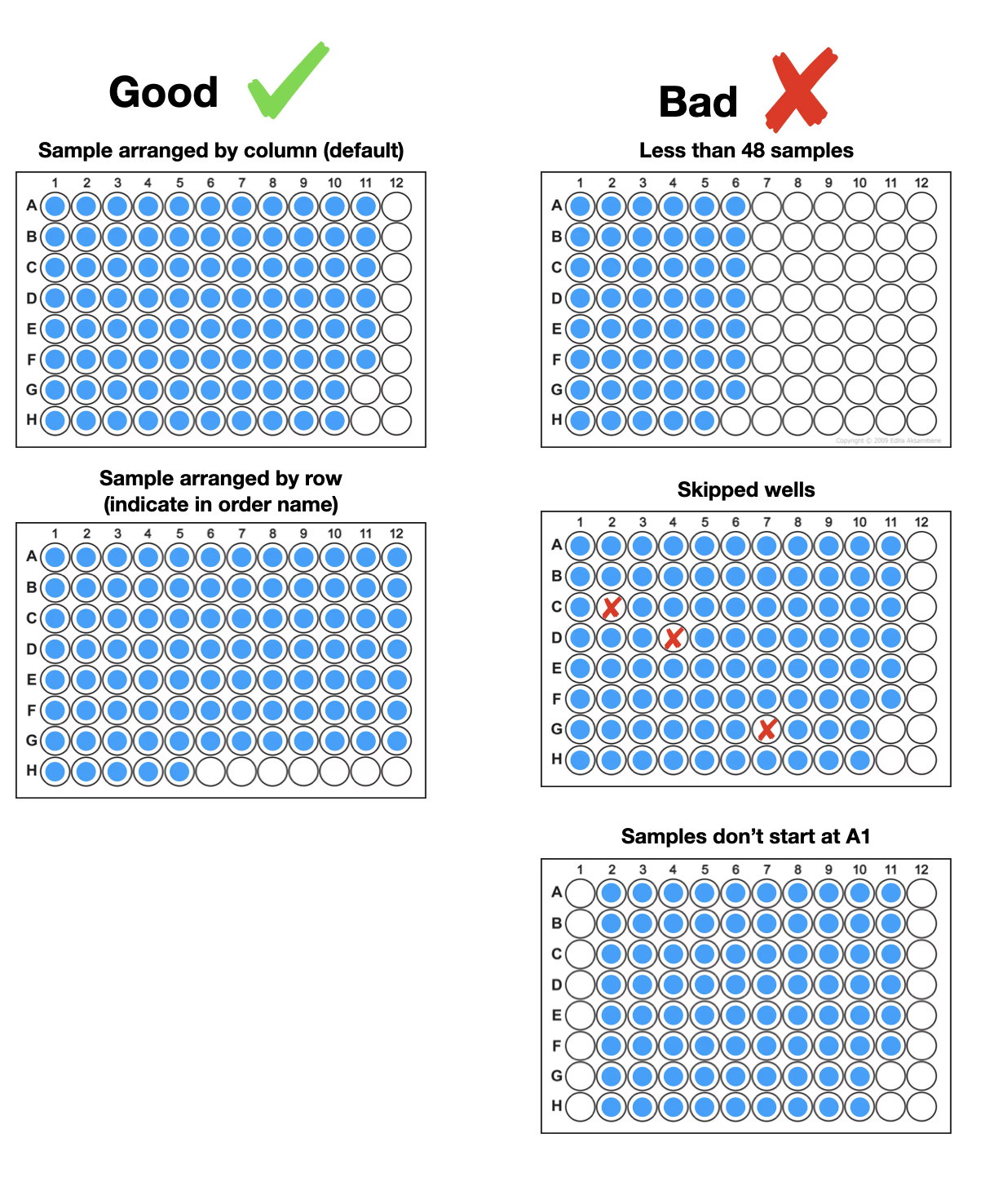
Start with well A1 and load your samples contiguously. Our preference is for samples to be loaded by column but we can accept samples loaded by row if indicated in the order. Do not leave any empty wells between samples.
- Ensure your plates are sealed extremely thoroughly to survive shipping without leaking — sadly we receive a lot of leaky plates. We recommend using a heat seal (rather than adhesive) or snugly-fitting strip caps on top of the plate. If you only have adhesive options, click here for some examples of sealing technique.
- To prevent cross contamination of samples, our customers have also had good success with dehydrating plates: spin the plate(s) down and then place the plate(s) on a 65 degree heating block for 0.5 to 1 hour or until dry. After dry, you can seal the plate with sticky film to prevent contamination of the sample during transport. We will rehydrate the sample in our lab for sequencing.
- Wrap each plate individually in bubble wrap or other packaging material to prevent cracking and seal punctures. DO NOT stack multiple plates directly on top of each other, as the wells of one plate will likely puncture the seal of the next plate.
Dropoff or ship.
Print out your order form. Fold so that QR code is readily visible and place in a small bag with your samples.
For fastest results and easiest shipping, place your samples in a Plasmidsaurus Dropbox!
If you ship your samples to us directly:
- Place packaged samples in a sturdy cardboard or padded envelope or a cardboard box. If reusing old packaging, ensure that any hazard warnings are completely obliterated. Multiple orders can be shipped in the same package, as long as each order is placed into a separate bag with its corresponding order confirmation sheet.
- Do not add ice packs or dry ice. Samples should be shipped at room temperature.
- Ship via any Express or First-Class shipping carrier such as FedEx, UPS, DHL, or USPS. The shipping address will be provided on your order confirmation sheet.
If you aren't using a Plasmidsaurus dropbox, print and fill out the following documents:
- Commercial Shipping Invoice - DNA
For best results, make sure to include the shipper's identity, shipper's address, and the quantity of items within the shipment.
- Detailed Description - DNA
- Toxic Substances Control Act (TSCA) Certificate - DNA (only if shipping from outside of US to US)
Further guidance for international shipping
- If shipping to Cologne, certain sample types will require some additional information. Please reach out to support if you have any questions.
- Make sure all forms are fully completed. Incomplete and/or missing forms will likely cause delays and other issues.
- Keep an eye out for any communication or information requests from your shipping company of choice in case of issues. It is your responsibility to provide any information they request, and we advise following their guidance as their systems will have the most up-to-date customs information.
- Samples must be packaged and shipped in accordance with U.S. Department of Transportation (DOT) and/or International Air Transport Association (IATA)/ International Civil Aviation Organization (ICAO) and carrier regulations.
- The information below constitutes only a recommendation for shipping samples classified as "non-regulated materials" to our facility. Due to continuing changes in state and federal regulations, clients should always check with their safety office and/or shipping department to ensure regulatory compliance.
- Your international courier will require you to complete a commercial invoice to be included with the shipment.
- The commercial invoice should describe the contents as “non-hazardous, non-contagious research sample.” Please list your institution as the "Manufacturer" of the samples if Manufacturers Identification (MID) code is required.
- State that the “contents are Not Restricted under IATA DGR regulations.”
- We recommend that you include a Toxic Substance Control Act (TSCA) Certification. If you do not include this, the package may be delayed at customs until the courier receives the proper certification.
- Declare the value of the goods to be $1.
- We only accept pure DNA samples that are intended for Research Use Only! We DO NOT accept any samples for clinical diagnostics or trials. Any samples that do not conform to our submission guidelines will be discarded and/or submitted to the United States Postal Inspection Service.
Why do I need these forms? Read more here.