Plasmidsaurus speed at genome scale
Most whole genome sequencing still relies on short reads, slow turnaround times, and fragmented assemblies that miss the bigger picture. Plasmidsaurus Whole Genome Sequencing lets you go straight from cells to complete genome assemblies in as little as a day, bypassing the need for tricky sample prep or complex bioinformatics pipelines. With Plasmidsaurus convenience and speed, you can discover new biology, design new pathways, and monitor your experiments with the full picture.
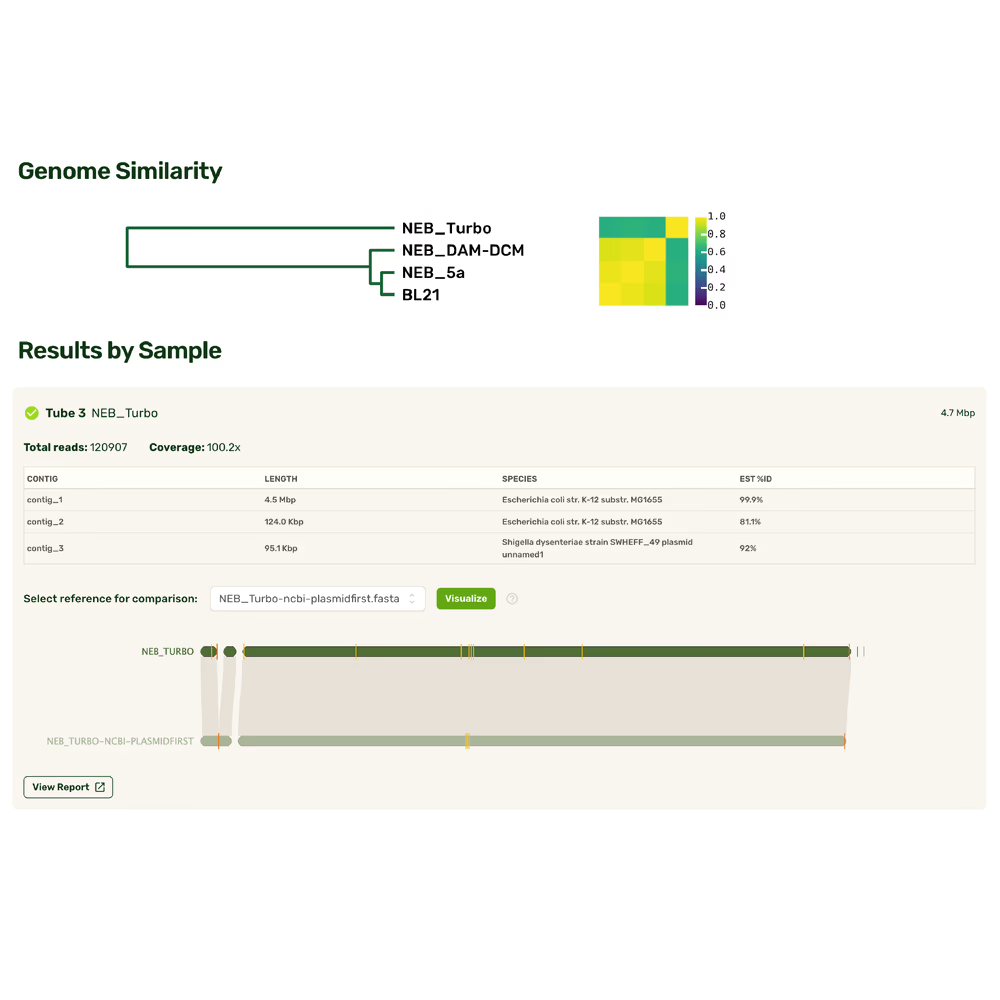
Fast answers, full coverage
Complete coverage
Achieve complete, gapless genome assemblies with de novo assembled long reads — capturing regions and structural features that short-read assemblies leave fragmented or unresolved
Maximal convenience
Start from cells and get assembled genomes. No need to master new sample prep or bioinformatics techniques—spend time interpreting results and leave the rest to us.
Rapid results
Get high-accuracy genome assemblies from extracted DNA within days.
Which organism are you working with?
Organism
Cultured bacterial and archaeal isolates with genomes up to 12 Mb
Sample input
Cell pellets preserved in Zymo DNA/RNA Shield™ or extracted gDNA
DNA extraction available for
All BSL1 and BSL2 strains
As fast as
1 - 2 business days.
See service level details below for more info.
As low as
$90
Organism
Cultured budding or fission yeast isolates with genomes up to 20 Mb
Sample input
Cell pellets preserved in Zymo DNA/RNA Shield™ or extracted gDNA
DNA extraction available for
18 genera of yeast
As fast as
1 - 2 business days
See service level details below for more info.
As low as
$150
Organism
Organisms with genomes from 20 Mb to 3.3 Gb including protozoans, plants, animals, and humans
Sample input
Cell pellets preserved in Zymo DNA/RNA Shield™ or extracted gDNA
DNA extraction available for
Most organisms and cell types without a cell wall, including human tissues
Questions? Contact us
As fast as
3 - 6 business days.
See service level details below for more info.
As low as
$250
Rethink your genome sequencing workflow
Bioprospecting
Get de novo assemblies of genomes from novel isolates or wild organisms for functional characterization. Discover new enzymes, proteins, or pathways with potential industrial or biomedical applications.
Directed evolution
Quickly and reliably identify complex mutations that lead to desirable phenotypes, including rearrangements, duplications, insertions, and integration events that short-read or targeted approaches often miss.
Strain engineering
Sequence periodically throughout a strain editing campaign to establish baseline truth, verify that intended genome edits were successful, identify off-target modifications, and validate prior to production.
Structural variant detection
Accurately detect a wide variety of structural variants that are missed by short read platforms.
Integration mapping
Characterize integration sites after genome editing, insertional mutagenesis, or retroviral infection
Need ultra-precise SNP detection? Choose hybrid!
Our optional Hybrid services supplement your Oxford Nanopore-based data with Illumina reads (2 x 150bp paired-end sequencing) for polishing errors in tricky homopolymers and methylated motifs. Our Illumina data targets (not guaranteed) are:
- Bacteria Hybrid: 100 Mb
- Bacteria Big Hybrid: 170 Mb
- Yeast Hybrid: 290 Mb
Not available for Eukaryotic Genome Sequencing.
"Nothing feels quite like "We live in the future" more than getting back sequenced and assembled bacterial genomes in less than 24 hr. Thanks Plasmidsaurus!"
Product specs & service levels
- Add $15 and 1-2 business days to turnaround time for DNA extraction
- We support extraction from a single isolate of any BSL1 or BSL2, gram + or - bacterial species.
| Service Level | Size | Sequencing Platform | Sample Submission from gDNA | Sample Submission from Cells | Cost | Target Turnaround Time |
|---|---|---|---|---|---|---|
| Standard | <7 Mb | ONT | 20 μL at 50 ng/μL normalized concentration | 4-6 x 10^9 cells in 500 µL Zymo DNA/RNA Shield™ | $90 | 1 business day |
| Big | 7-12 Mb | $105 | ||||
| Hybrid | <7 Mb | ONT + Illumina | 30 μL at 50 ng/μL normalized concentration | $165 | 6-8 business days | |
| Big Hybrid | 7-12 Mb | $195 |
- Add $15 and 1-2 business days to turnaround time for DNA extraction
- We support extraction from the following host types: Asbya, Kluyveromyces, Candida, Lipomyces, Debaryomyces, Metschikowia, Eremothecium, Pichia/Komagataella, Endomyces, Saccharomyces, Hansenula, Saccharomycopsis, Hanseniaspora, Schizosaccharomyces, Issatchenkia, Torulopsis, Kloekera, Yarrowia.
| Service Level | Size | Sequencing Platform | Sample Submission from gDNA | Sample Submission from Cells | Cost | Target Turnaround Time |
|---|---|---|---|---|---|---|
| Standard | <20 Mb | ONT | 20 μL at 50 ng/μL normalized concentration | 50 - 100 mg cell pellet in 500 µL Zymo DNA/RNA Shield™ | $150 | 1 business day |
| Hybrid | <20 Mb | ONT + Illumina | 30 μL at 50 ng/μL normalized concentration | $255 | 6-8 business days |
- Add $50 and 2-3 business days for DNA extraction. We support DNA extraction from human and mouse tissues, as well as most organisms and cell types that lack a cell wall. A minimum of 1 × 10⁶ cells is required. Question about whether we can extract your sample? Contact Support.
- How do I determine my data target? Read this FAQ here!
- If you have fewer cells available than we require in the table below, we can often work with lower inputs too - please email us at support@plasmidsaurus.com to discuss options.
- De novo assembly and annotation not guaranteed and depends on sample complexity.
| Service Level | Genome Size Range (30X Coverage) | Sequencing Platform | Sample Submission from gDNA | Sample Submission from Cells | Cost | Target Turnaround Time |
|---|---|---|---|---|---|---|
| 1 Gb data target | 20-60 Mb | ONT | 20 μL at 50 ng/μL normalized concentration | 1 x 10^6 cells in 200 µL Zymo DNA/RNA Shield™ | $250 | 3-5 business days |
| 5 Gb data target | 75-250 Mb | 40 μL at 50 ng/μL normalized concentration | $500 | |||
| 15 Gb data target | 300-750 Mb | 60 μL at 50 ng/μL normalized concentration | 2 x 10^6 cells in 200 µL Zymo DNA/RNA Shield™ | $1000 | 5-8 business days for FASTQ data; 1-4 weeks for assemblies | |
| Full flow cell (50-100 Gb data target) | 1 - 3.3 Gb | 60 μL at 50 ng/μL normalized concentration | $1750 |
Looking for more data? Working with an organism with a bigger genome?
Check out our custom sequencing solutions to see how we can help.
Data deliverables & bioinformatics
Genome reference assembly
Functional annotation
Assembly statistics
Sequencing statistics
Raw reads
Assembled contig ID (for bacteria only)
Interactive genome alignment (for bacteria only)
Ready to sequence?
Send us extracted and purified genomic DNA or cell pellets suspended in Zymo DNA/RNA Shield™. We'll handle the rest.
Relevant resources
Related products
Microbiome
Got a mix of species? Check out Microbiome Sequencing for 16S, 18S, or ITS sequencing of your community.
Amplicon
Need a more targeted approach? Check out Amplicon Sequencing.
Custom
Is your genome giant? Do you need full assemblies of all species in your community? Work with us through Custom Sequencing.
FAQs
Plasmidsaurus Whole Genome Sequencing is performed using the newest long-read sequencing technology from Oxford Nanopore Technologies (ONT).
We use a transposome complex to construct an amplification-free long-read sequencing library, including minimal fragmentation of the input genomic DNA in a sequence-independent manner, and return you the annotated genome assembly. For more details on our workflows and bioinformatics pipelines, please see technical documentation for Whole Genome Sequencing.
Our bacterial, yeast, and eukaryotic services offer similar library preparation and sequencing quality but specialized genome extraction services, sequencing depth, and bioinformatics pipelines optimized for bacteria, yeast, and eukaryotes (including filamentous fungi and multicellular animals).
We recommend choosing the option that matches the organism you are sequencing. If you need more read depth for yeast sequencing, you can submit yeast samples to the Eukaryotic genome services that offer higher data targets. Deeper sequencing is also available for multicellular genomes via our Custom Service.
The Hybrid ONT + Illumina option for Bacteria and Yeast Genome Sequencing polishes your long-read ONT assembly with Illumina short reads (paired-end 2x150bp) using the Polypolish v0.6 tool to improve the consensus quality at those tricky homopolymer runs and methylation motifs.
This Hybrid service is ideal when:
- You anticipate an abundance of homopolymers and/or methylated motifs in the genome
- You are planning to use the genome assembly result as a reference for SNP variant calling
The Hybrid service is not generally necessary if your primary goal is to obtain a high quality, high coverage, highly contiguous genome assembly, as contiguity is entirely determined by the quality and quantity of the ONT long read data. Adding in short Illumina reads with the Hybrid service has no impact on the number, size, or topology of assembled chromosomes/elements, and instead only improves nucleotide-level basecalling accuracy in homopolymers and methylated motifs via polishing. For this reason, we do not repeat the genome assembly or contig identification workflows after Illumina polishing.
Please note: Hybrid ordering options are not currently available for the Eukaryotic Genome, but you can request to add Illumina data to your ONT eukaryotic data through the Custom Sequencing service.
The Eukaryotic Genome Sequencing service offers four different service levels, which allow you to select the target amount of output per sample, measured in Gb of fastq data. We don't specify output in number of reads because that varies based on the specific read length distribution in question.
The Service Level recommendations below are based on a target of approximately 30x genome coverage, which is typically sufficient to generate a high-quality de novo genome assembly with annotations. Depending on your application, you can submit your sample to a different service level than is recommended in order to obtain a different amount of coverage. For example:
- If you plan to use the raw reads to perform copy number variant analysis against a reference genome, you might need less genome coverage, e.g. 10-20x. Please note that while the assembly is more likely to fail at lower coverage levels, you will still be able to use the raw reads to perform your analysis
- If you plan to use the raw reads to generate a haplotype-phased de novo genome assembly, in some cases you might need more genome coverage, e.g. 60-100x
| Service Level | Example Species | Approx. Genome Size (Haploid) | Approx. Genome Coverage |
|---|---|---|---|
| 1 Gb | Plasmodium falciparum (protozoan) | 23 Mb | 43x |
| Fusarium oxysporum (fungus) | 36 Mb | 28x | |
| Neurospora crassa (fungus) | 43 Mb | 23x | |
| 5 Gb | Caenorhabditis elegans (nematode) | 100 Mb | 50x |
| Daphnia pulex (water flea) | 125 Mb | 40x | |
| Drosophila melanogaster (fruit fly) | 180Mb | 28x | |
| 15 Gb | Anopheles gambiae (mosquito) | 278 Mb | 54x |
| Oryza sativa (rice) | 430 Mb | 35x | |
| Mimulus guttatus (monkeyflower) | 450 Mb | 30x | |
| 50 - 100 Gb (1 flow cell) | Danio rerio (zebrafish) | 1.5 Gb | 33-66x |
| Mus musculus (mouse) | 2.7 Gb | 18-36x | |
| Homo sapiens (human) | 3.3 Gb | 15-30x |
The typical turnaround time in business days is listed below for each service. This is the average time it takes to return data to customers who are submitting orders from US, UK and EU. Turnaround times for customers in APAC may be slightly longer due to the time required in transit to our lab in Singapore.
Low sample quality can also negatively impact turnaround time and the quality of your results. Please read sample prep instructions carefully to make sure your samples meet input requirements. Due to variability in shipping logistics and sample quality that is outside our control we cannot guarantee turnaround times or sample success rates.
| Service | From pre-extracted gDNA | With extraction | Hybrid from pre-extracted gDNA | Hybrid with extraction |
| Bacteria or yeast | 1 | 2 | 6-8 | 8-10 |
| Eukaryotic 1 Gb & 5 Gb | 3-5 | 5-8 | N/A | N/A |
| Eukaryotic 15 Gb & Full flow cell | 5-8 days for FASTQ data 1-4 weeks for assemblies | 8-11 days for FASTQ data 1-4 weeks for assemblies | N/A | N/A |