Ultrafast Transcriptomics with Plasmidsaurus RNA-Seq

RNA sequencing opens a window into the active life of cells. By providing a snapshot of the transcriptome at specific times and in different contexts, researchers can connect genetic information to real-time cellular behavior to uncover disease signatures, track how cells respond to therapies, or explore new pathways in biotechnology. Despite its transformative potential, RNA-seq has remained difficult for many labs to access: too expensive, time consuming, and requiring specialized technical knowledge and complex data analysis.
At Plasmidsaurus, we are obsessed with making sequencing services that fit into the flow of experimental science, eliminating processes that add hassle to your day so you can focus on your science. We pioneered Whole Plasmid Sequencing to give scientists insight into what was happening in their cloning workflows, with easy setup up, fast sequencing, and instant insights with our bioinformatics tools. Today we’re so excited to launch our first RNA-Seq service, making unbelievably fast and hassle-free gene expression analysis available.
Plasmidsaurus RNA-Seq offers ultra-fast genome-wide gene expression analysis at the speed of qPCR. Just send us cells preserved in Zymo DNA/RNA Shield™ and get full transcriptome analysis. The process is simple:
- Drop cells preserved in Zymo DNA/RNA Shield™ in a Plasmidsaurus dropbox or ship them to our lab (you can order Zymo DNA/RNA Shield™ using the checkout code PLASMIDSAURUS for 10% off!)
- Wait 3 days if you’re in the continental US (or one week globally) while we prep your library and run short read sequencing
- Get up to 10 million deduplicated reads per sample and intuitive, interactive results that reveal the state of your cells
How it works
For sequencing library preparation, we convert mRNA into complementary DNA (cDNA) via reverse transcription and second-strand synthesis, followed by tagmentation and library indexing and amplification. We then sequence the libraries using Illumina NovaSeq instruments, generating an extraordinary number of unique reads per sample.
Our first RNA-Seq product uses 3’ end counting to capture differential gene expression. This method allows for extremely efficient and accurate counts of mRNA transcripts. With other methods of RNA-seq, full length cDNA molecules are fragmented and multiple parts of the same cDNA transcript are sequenced. Typical transcripts are about ~1-2 kb, with some being much longer, meaning many 150 bp reads are needed to span each molecule. As a result, not every new read is biologically informative, limiting the efficiency with which sequencer capacity is used. Additionally, longer transcripts require more reads, leading to a length bias in the data analysis that can artificially give longer transcripts greater statistical power than shorter ones (Oshlack & Wakefield, 2009).

3’ end counting allows us to more efficiently profile the transcriptome by sequencing only from the 3’ end (Figure 1). This approach produces reliable gene expression levels comparable to full-length methods (Alpern et al., 2019), while needing far fewer reads, and also being relatively resistant to RNA degradation. Additionally, the number of reads is directly related to the number of transcripts of a given gene, and there will be no difference in coverage of longer and shorter transcripts. When combined with Plasmidsaurus’ industry leading turnaround times, Plasmidsaurus RNA-Seq is able to deliver insights into the genome-wide real-time state of your study system in record time (Figure 2).
Why Plasmidsaurus?
Traditional transcriptomics studies are a huge commitment. They’re generally slow, expensive, and require a large minimum number of samples. We believe that if scientists could get transcriptome-wide insights in days not weeks, very easily and affordably—without minimum order sizes or complicated vendor onboarding—it will enable a dynamic and rapidly iterative approach to cell biology and research and engineering that’s never really been possible before. Validate transfectants, the results of your CRISPR edits, or stem cell differentiation in days, with a transcriptome-wide view. Rapidly screen drug treatments for impact on target pathways. Or even just monitor the health and stability of your cultures.
Our veritable army of galaxy-brained R&D scientists spent many months assessing the various approaches to cDNA sequencing, evaluating the conversion efficiency, reproducibility, and accurate assessment of input abundance. Their meticulous work led us to what we believe will be a product that delivers unmatched insights at an unprecedented price.
To balance speed, cost, and statistical power, we are providing 10M deduplicated 3’ end counting reads per sample. Our aim is to have a turnaround time of three days, though it may take us a short while to optimize enough to deliver this lofty target every time. And just to emphasize in case you weren’t aware: three days is approximately ten times faster than the industry standard of three weeks.
Sample input and preparation guidelines
RNA extractions and library preparation is a hassle, and can be prone to human error. With Plasmidsaurus, we handle the RNA extraction and library prep for you! Let our automation handle the less exciting stuff, so that you can get on with your experiments.
You also don’t have to worry about cold chain. Simply submit ~100,000 cells in 50 μL of Zymo DNA/RNA Shield™, either through our extensive network of drop boxes, or via direct shipping, at your preference. Our team of customer support scientists is available to help you should you have any questions or concerns around sample preparation and shipping. See our detailed sample prep instructions for more information.
Cutting-edge differential gene expression analysis delivered right to your inbox
At Plasmidsaurus, our goal is to supply you with actionable biological insights, saving you from the hassle of installing tools, maintaining databases, and debating the merits of random subsampling versus negative binomial approaches to dealing with uneven sample depth.
Your output reads will come piping hot off the sequencer and will be run through a rigorous set of QC steps: filtering, trimming, and all the rest. Our DGE pipeline will align your reads to your chosen species’ reference genome, inclusive of any exogenous sequences (e.g. viral or transgene) you’d like to include. On the results page you can easily group samples by relevant treatment conditions and launch pairwise differential gene expression analyses.
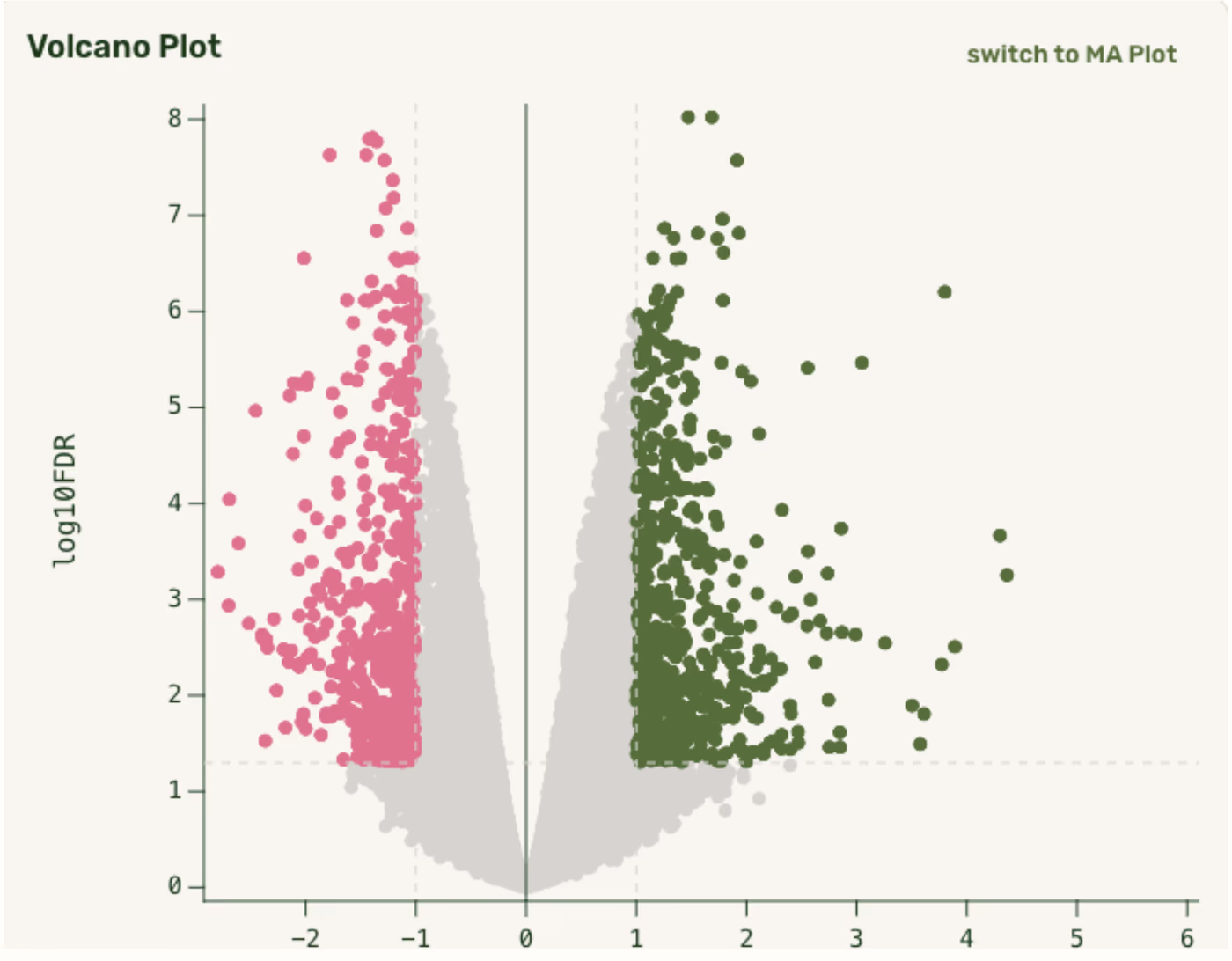

Within a few seconds you’ll have common DGE plots, such as volcano plots, gene bar plots, and pathway enrichment plots, that you allow you to interactively explore your data. If you want to dive deeper, we’ll also provide you with aligned deduplicated bam files, raw FASTQ files, tabular gene counts and sequencing run level QC information.
What does the future hold?
In typical Plasmidsaurus style, we’ll continue to add analyses and visualization tools to the results page, and you’ll always be able to apply these new tools to your legacy orders. Beyond DGE, we know a lot of you would like to see a nanopore based full-length cDNA product, or maybe even a direct RNA product. We’re actively refining our long term product roadmap, so stay tuned and watch this space!
Ready to sequence? Check out the RNA-Seq page to get started.