RNA-Seq
Genome-scale insight at the speed of qPCR. Send cells and get analyzed transcriptomes in as fast as 3 days.

Ultrafast genome-wide gene expression analysis.
Traditional transcriptomics studies are slow, expensive, and require a large minimum number of samples. With Plasmidsaurus RNA-seq, get transcriptome-wide insights in days, not weeks, and transform your research with speed and insights that have never been possible before. Validate transfectants, CRISPR edits, or stem cell differentiation, rapidly screen drug treatments for impact on target pathways, or monitor the health and stability of your cultures.
Fast turnaround, simple setup, comprehensive data
Sample input
Cultured animal cells lysed in Zymo DNA/RNA Shield™ or purified eukaryotic RNA. See sample prep for important details.
Shipping
For cells: ambient temp shipping direct to our lab, or use our dropbox network
For purified RNA: ship on dry ice to our lab
Data deliverables
Up to 10M deduplicated Illumina reads, count tables, and Run QC information
As fast as
Less than 3 days if shipped within the US
~ 1 week outside the US
Price per sample
$50 for academia
$80 for industry
Unmatched speed
Go from crude cell preps to fully analyzed data in as fast as 3 days, 10x faster than industry average.
Genome-wide insight
Measure expression of all polyadenylated transcripts at costs competitive with targeted methods like qPCR.
No bioinformatician needed
Explore gene expression changes without needing to be a bioinformatician. Leave that to us!
No library prep, no cold chain
Harvest cells in Zymo DNA/RNA Shield™ and place them in your nearest dropbox.
Unbiased data
No transcript length bias. The number of reads is directly related to number of transcripts.
Affordability without compromise
Premium sequencing accuracy and data quality at dramatically lower cost than other options.
"This is SO COOL!!!!! Huge kudos to Plasmidsaurus for this incredibly fast, affordable service. We'll definitely love using this service in the future."
Ideally suited for a range of applications
Drug Development
Pharmaceutical and biotech companies seeking rapid feedback on drug candidates or genome editing.
Genome-Scale Research
Labs studying gene regulation, cell differentiation, genome engineering, and developmental processes.
Cell Culture
Anyone who needs rapid validation of cell line identity and health.
Sequencing moves fast. We stay on top of it for you, keeping you at the bleeding edge.
We use the right technology for the problem, partnering with leading sequencing technology providers for early access to the latest chemistries, hardware, and bioinformatics.

Interactive & intuitive results
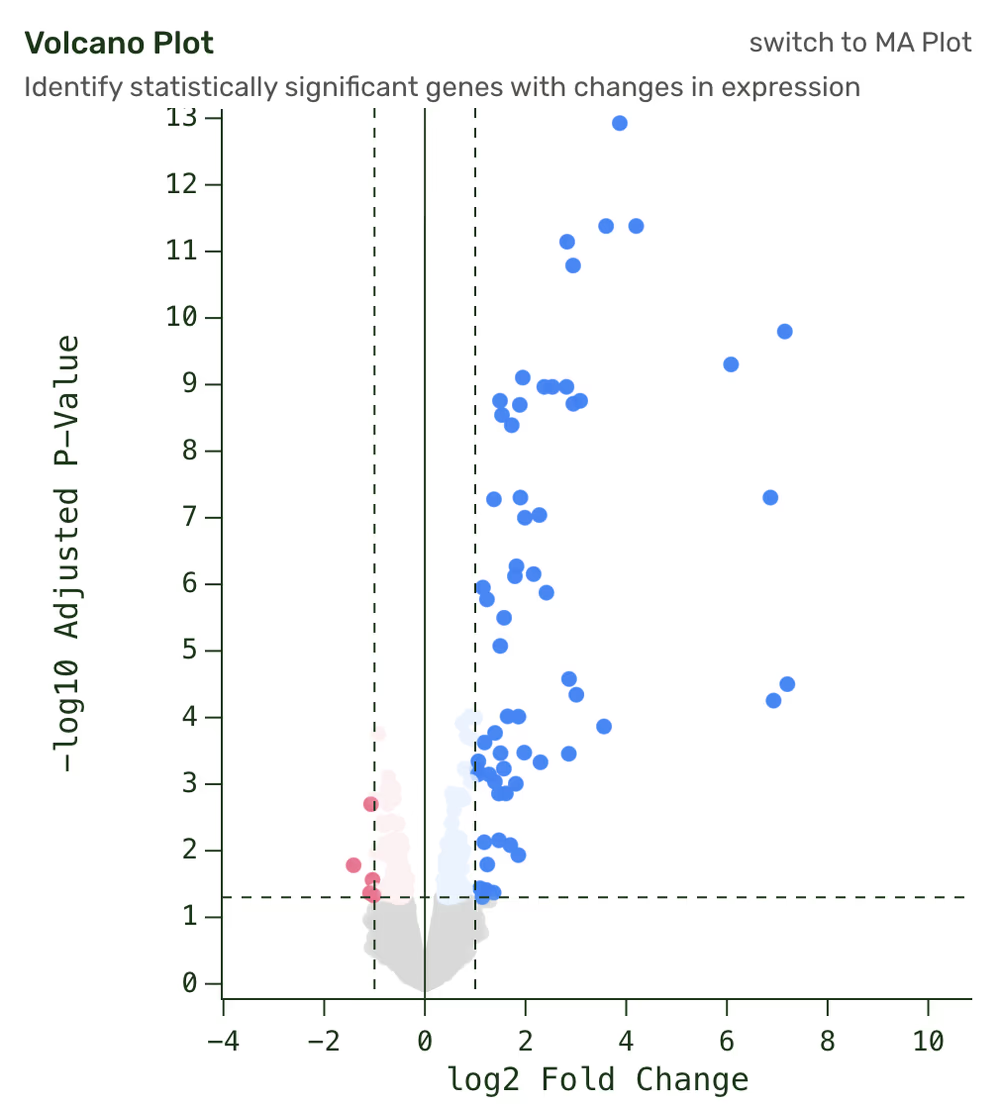
Interactive volcano plot
Visualize differential gene expression patterns of up- and downregulation.
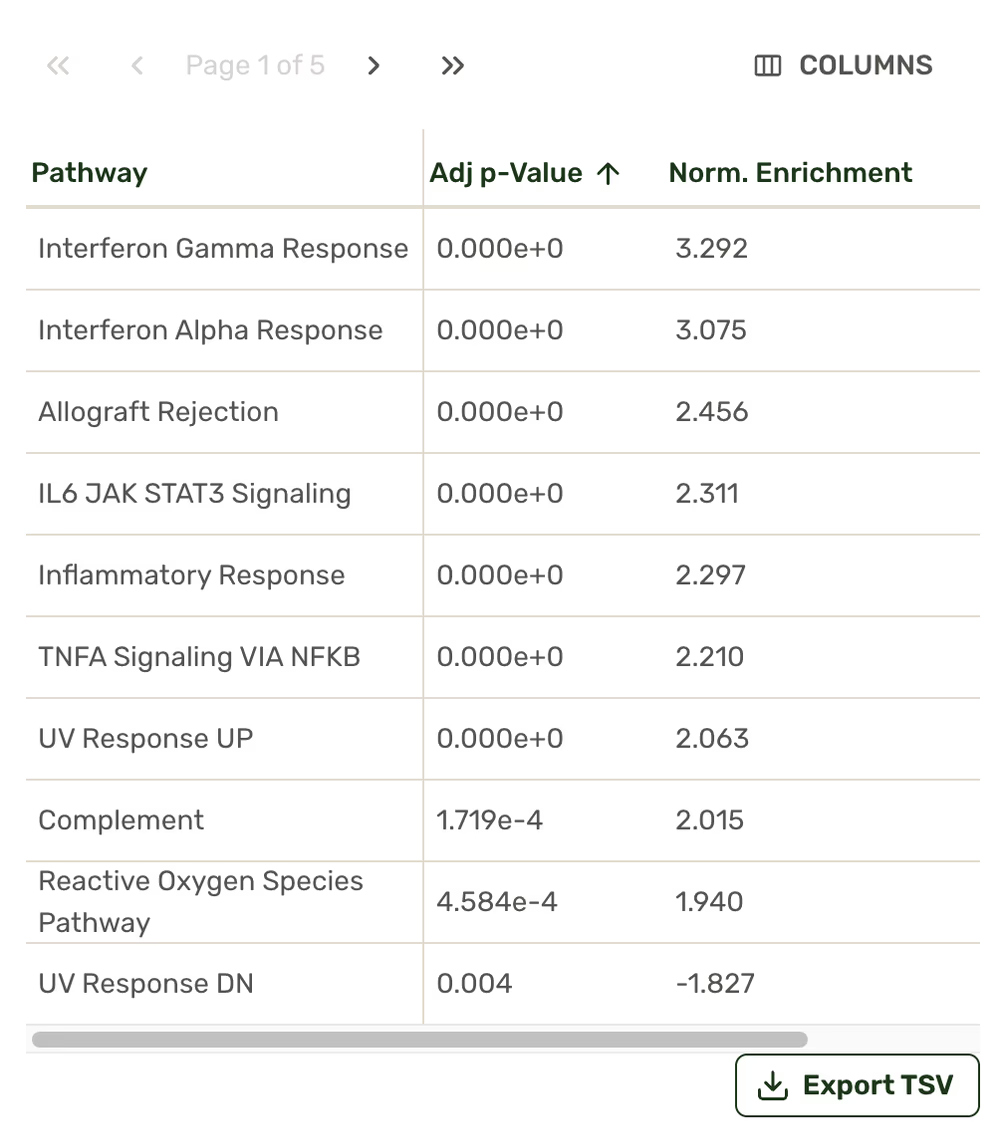
Functional enrichment plot
Track the most affected regulatory pathways in your samples.
Per-sample expression profiles
Select genes or pathways you want to analyze and see them visualized.
Try out our live demo with real data
Other data types
Compare expression levels and explore highly enriched functional pathways.
Interactive gene expression profiles

Interactive volcano plots
Functional enrichment plots

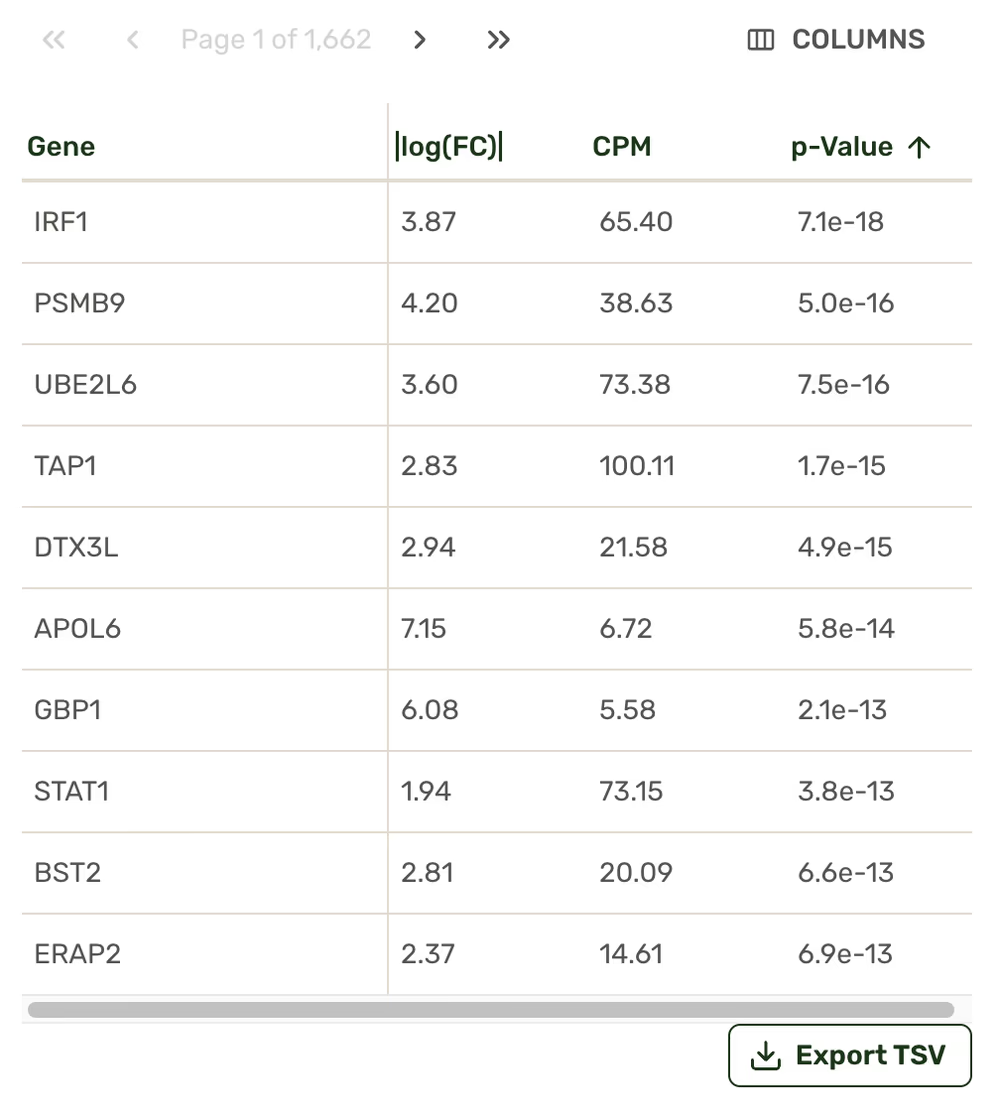
Gene count tables
Enriched pathway tables
Deduplicated, aligned .BAM files
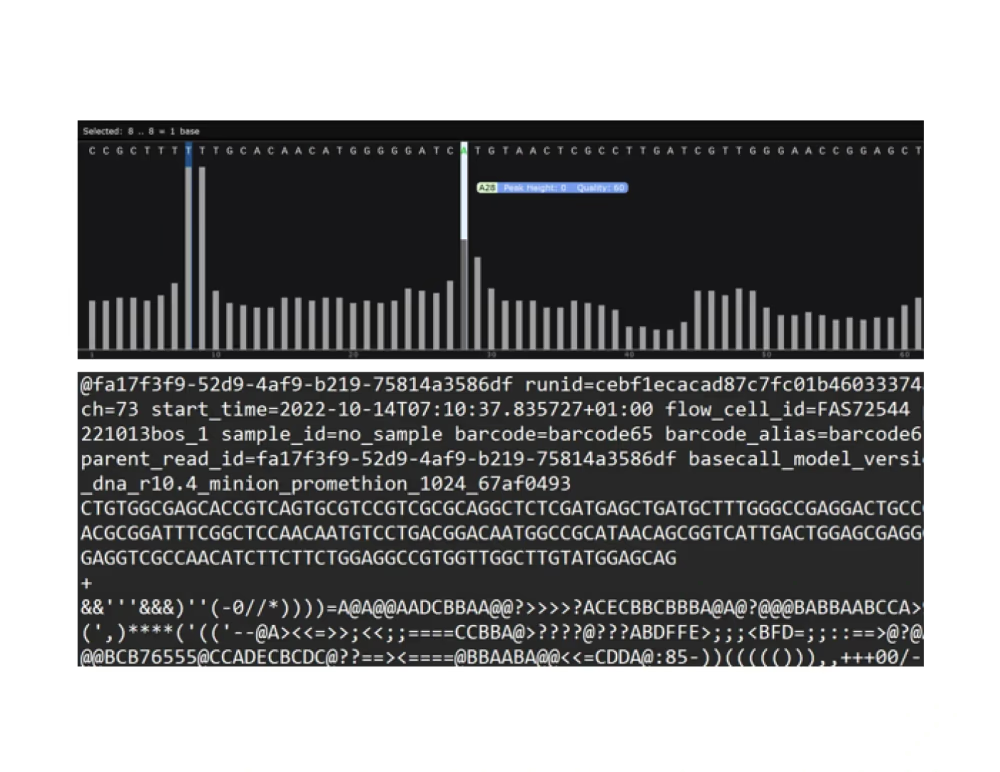
Raw FASTQ reads

Sample correlation matrix

Per-sample gene counts (.TSV)

Quality control summary (fastqc)

Ready to sequence?
Submit ~100,000 cultured cells in Zymo DNA/RNA Shield™ at room temperature. We'll handle the rest.
Relevant resources
FAQs
Plasmidsaurus RNA-Seq provides 3’ end counting for transcriptome-wide differential gene expression data. We utilize the Illumina platform to provide ~10M deduplicated 3' end counting reads (20M raw reads) from ~100k cells or 300 ng purified RNA, returned in ~3 days.
Every order includes interactive data analysis tools on the results page. These tools enable immediate insights into your experiments, providing actionable data analysis within minutes of getting your results.
There is no minimum order size or quote process. Cell samples do not require cold chain shipping.
Our interactive results page lets you explore the most differentially represented genes across your samples, create and explore differential gene expression analyses (think Volcano Plots), detect enriched cellular pathways, select genes to track across conditions, and more. Pathway and gene count comparisons are all exportable.
If you’d like to do your own offline analysis, we provide the following downloadable content: quality control summary (fastqc), aligned deduplicated bam files, and gene counts in structured plain text format. You can also download the raw fastqs.
The goal of Plasmidsaurus RNA-Seq is to accurately quantify transcript abundance. Because PCR creates copies, we use unique molecular identifiers (UMIs), which are added during cDNA creation. This allows us to differentiate between a multitude of unique mRNA molecules versus a molecule that was amplified.
Clustering by UMI sequence allows us to analyze and return deduplicated reads only: this is the most accurate way of quantifying transcript abundance.
The gold standard method to determine the accuracy of any RNA-Seq method is to spike in 92 synthetic RNA sequences at known abundance, designed by the consortium for RNA sequencing (ERCC). We spiked those sequences into 100 ng of universal human reference RNA at a standardized concentration, and then performed Plasmidsaurus RNAseq. Plotting the true concentration against measured abundance reveals that we can accurately quantify true transcript abundance across multiple orders of magnitude of RNA abundance, down to at least 1 CPM.
Library preparation contains the following steps:
- Poly-A mRNA capture and reverse transcription for cDNA synthesis: oligo-dT primer to capture just the mRNA (contains sample barcode, UMI, and Read 1 sequences).
- Second strand synthesis and tagmentation: double stranded-cDNA is generated. Tagmentation creates fragments and incorporates Read 2 sequence.
- Illumina library amplification: adds unique dual indices (UDI = i5, i7) and P5/P7 sequences.
At this time, we are accepting the following sample types:
- Preserved animal cells and plant protoplasts: This includes adherent, suspension, and primary cell cultures. For best results, cells should be healthy, actively growing, and free from contamination.
Purified RNA: We can accept purified bulk RNA, free from DNA and RNAse activity, shipped with dry ice. We need at least 30μl of at least 10ng/μl concentration.
We are currently NOT accepting the following sample types:
- Prokaryotic samples of any type.
- Yeast and other fungal cells (purified RNA is acceptable.)
- Tissue Samples and organoids: We are unable to process tissue samples at this time, but we look forward to offering this in the future. Extracted RNA is a great alternative.
Please send ~100k cells in 50 μL of 1X Zymo DNA/RNA Shield™, either through our extensive network of drop boxes, or via direct shipping at room temperature—no cold chain required. If you’re submitting purified RNA, we cannot provide free shipping at this time.
For all the details on sample input guidance, please see here.

