Rethink your microbiome workflow
Most microbiome studies are still built around large batch sequencing runs with slow turnaround times and workflows that rely on older short-read technologies. Plasmidsaurus Microbiome Sequencing offers a new model: full-length 16S, 18S, or ITS reads with more precise species-level classification, delivered as quickly as overnight.
Designed for speed, precision, and flexibility, Plasmidsaurus microbiome sequencing services are ideal for monitoring environmental and synthetic communities, understanding an experimental biome and how it responds to stimuli, and finding new species in wild communities. Send us raw biological samples, genomic DNA, or purified amplicons and quickly receive full-length reads and species-level classification.
Confirm what's in your community
Confirm your system has the expected species diversity before starting your experiment, with insights direct from raw samples, genomic DNA, or PCR amplicons.
Monitor with rapid results
Easily monitor your microbial communities over time, across locations, and in response to stimuli, with full length reads with taxonomic classification in under one day.
Discover with species-level clarity
Precisely identify microbial species that short-read sequencing can’t resolve. Discover what species are present and in what relative abundances.
We’re moving faster!
We now deliver Microbiome Amplicon sequencing results overnight for samples in Seattle, Los Angeles, and San Diego!
Which sample input is right for me?
Sample input

Raw samples (including soil, feces, swabs, and more)
Ideal for
Exploring the bacteria and archaea composition of a wide variety of samples
Service levels
Standard: Up to 5,000 reads
Big: Up to 10,000 reads
Huge: Up to 20,000 reads
Bronto: Up to 500,000 reads
As fast as
1 - 3 days
Availability
Within the US
As low as
$60
Sample input
Extracted environmental DNA
Ideal for
Samples requiring specialized extraction protocols that we don’t support or labs based outside the US
Service levels
Standard: Up to 5,000 reads
Big: Up to 10,000 reads
Huge: Up to 20,000 reads
Bronto: Up to 500,000 reads
As fast as
Next day
Availability
Globally
As low as
$45
Sample input

Purified amplicons of full-length 16S, 18S or ITS genes, or a mixture thereof
Ideal for
Studying fungi, protists, algae or other microbes that lack 16S genes and are commonly found in soil and water samples
Service levels
Standard: Up to 5,000 reads
Big: Up to 10,000 reads
Huge: Up to 20,000 reads
Add cleanup for $5
As fast as
Next day
Availability
Globally
As low as
$30
Product specs & service levels
All samples are sequenced with ONT.
| Service Level | Sample Input | Read Depth/Sample | Cost | Target Turnaround Time |
|---|---|---|---|---|
| Standard | Environmental samples | Up to 5,000 | $60 | 3 business days |
| Big | Up to 10,000 | $90 | ||
| Huge | Up to 20,000 | $150 | ||
| Bronto | Up to 500,000 | $265 |
All samples are sequenced with ONT.
| Service Level | Sample Input | Read Depth/Sample | Minimum Volume | Concentration | Cost | Target Turnaround Time |
|---|---|---|---|---|---|---|
| Standard | gDNA | Up to 5,000 | 10 µL min | 10 ng/µL | $45 | 1 business day |
| Big | Up to 10,000 | 20 µL min | $75 | |||
| Huge | Up to 20,000 | 40 µL min | $135 | |||
| Bronto | Up to 500,000 | 40 µL min | $250 |
All samples are sequenced with ONT. Add cleanup for $5 while placing your order. Cleanup adds 2 days to expected turnaround times.
| Service Level | Sample Input | Read Depth/Sample | Minimum Volume | Concentration | Cost | Target Turnaround Time |
|---|---|---|---|---|---|---|
| Standard | Linear double-stranded DNA | Up to 5,000 | 10 µL min | 10-20 ng/µL | $30 | 1 business day |
| Big | Up to 10,000 | 20 µL min | $60 | |||
| Huge | Up to 20,000 | 40 µL min | $120 |
Data deliverables & bioinformatics
Species and genus diversity
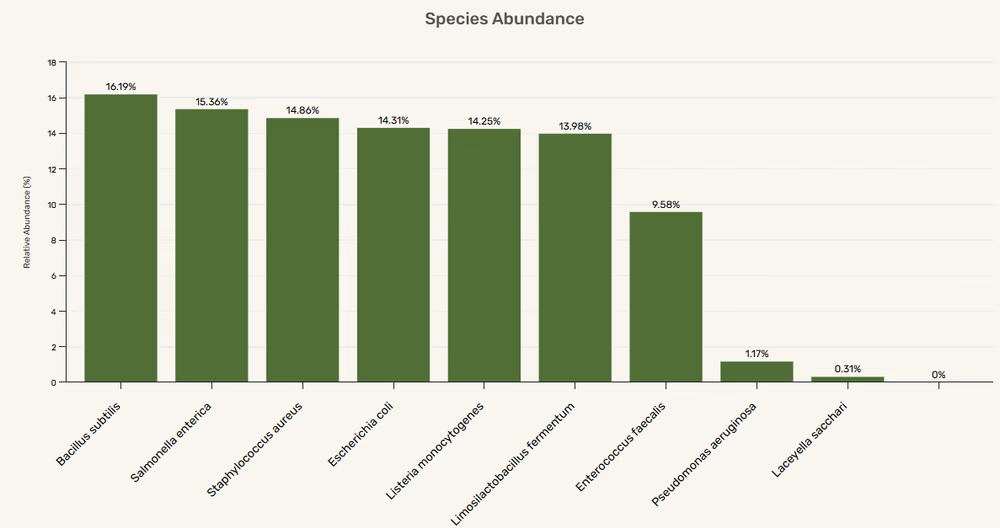
Taxonomic diversity
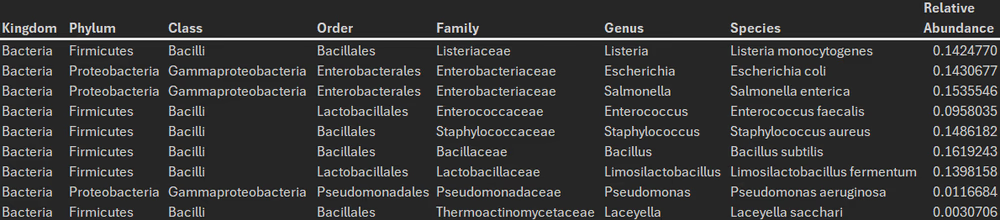
Read identifications
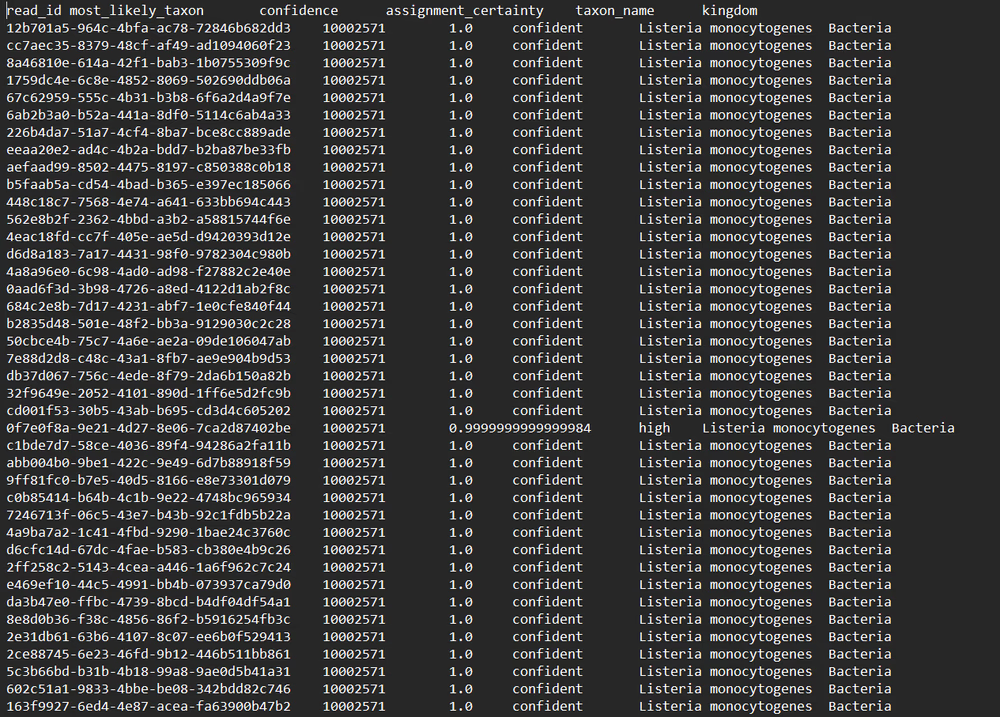
Statistics
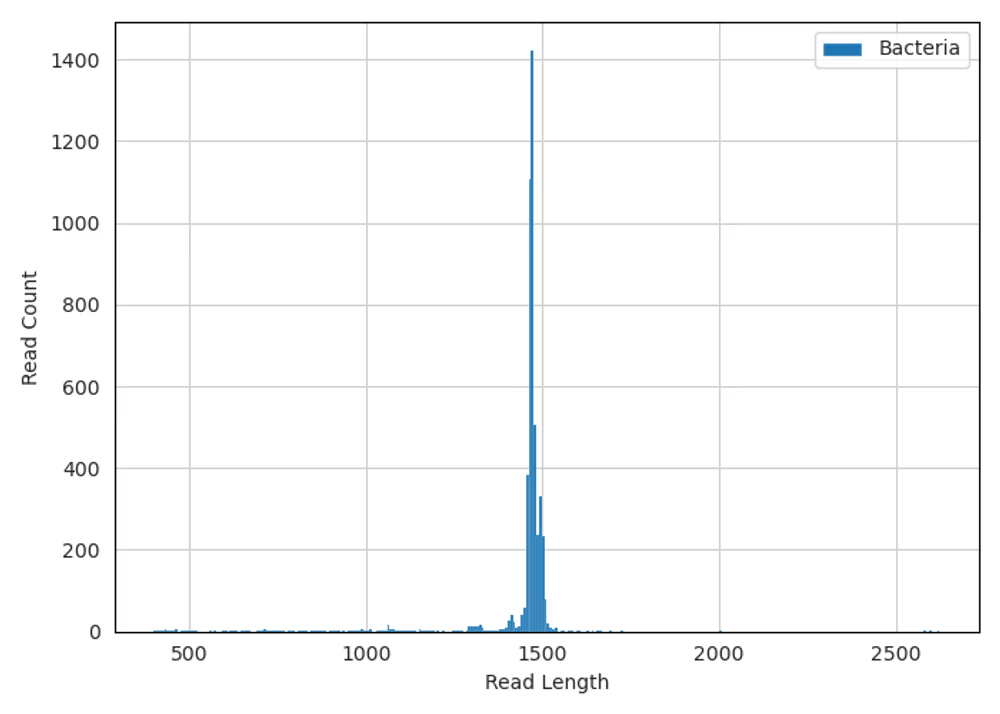
Raw reads
Ready to sequence?
Send your messy sample, purified DNA, or PCR amplicon. We'll handle the rest.
Relevant resources
Related products
FAQs
We sequence each sample the newest long-read sequencing technology from Oxford Nanopore Technologies (ONT), including the following components:
- Constructing long-read sequencing library using the newest v14 library prep chemistry.
- For the Microbiome 16S Amplification & Sequencing service, this includes barcoded full-length amplification of the 16S gene with our in-house sequencing primers.
- For the Microbiome Amplicon Sequencing service, this includes end-ligation of your 16S, 18S, and/or ITS amplicons. We do not further amplify your amplicons.
- Sequencing the library with a primer-free protocol using the most accurate R10.4.1 flow cells (raw data is delivered in .fastq.gz format), producing a full-length sequencing read for each amplicon.
As per Oxford Nanopore's specs for the chemistry and flowcells we currently use for Microbiome Sequencing, the raw read accuracy is typically >99.5%. We basecall the raw reads using ONT’s super-accurate basecalling model. We require a minimum raw read Qscore of 10 (90% accuracy) during sequencing.
We generate metrics on the relative abundance of all species that comprise at least 0.1% of the microbial community.
Zymo DNA/RNA Shield™-preserved source materials for extraction cannot be shipped via your local US dropbox, but instead must be shipped directly to our Eugene lab. You can request a free shipping label during order submission or ship on your own to:
Plasmidsaurus
1850 Millrace Drive, Suite 200
Eugene, OR 97403
If you would like us to attempt metagenomic DNA extraction from other types of materials that are not listed here, please contact us at support@plasmidsaurus.com to discuss options before order submission.